All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the Lymphoma Coalition.
The Lymphoma Hub uses cookies on this website. They help us give you the best online experience. By continuing to use our website without changing your cookie settings, you agree to our use of cookies in accordance with our updated Cookie Policy
Introducing

Now you can personalise
your Lymphoma Hub experience!
Bookmark content to read later
Select your specific areas of interest
View content recommended for you
Find out moreThe Lymphoma Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the Lymphoma Hub cannot guarantee the accuracy of translated content. The Lymphoma Hub and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
iwCLL 2017 | Epigenetics of CLL: understanding ontogeny and clinical behavior
Bookmark this article
At iwCLL 2017, on Monday 13th May, the second half of the session titled “Unraveling the factors leading to the development of CLL”, and was chaired by Emili Montserrat (Hospital Clinic of Barcelona) and John Gribben (Barts Cancer Institute).
During this session, a talk titled “Harnessing epigenetics to further our understanding of CLL ontogeny and clinical behavior” was presented by Christopher Oakes, PhD, from The Ohio State University Wexner Medical Center, Columbus, Ohio, US.
Oakes began the talk by explaining what epigenetics is: heritable information not encoded in the DNA sequence (A/T/C/G). Epigenetic states are programmed in cell lineage development and become fixed in mature, differentiated cells defining their cellular identity. Cancer cells acquire aberrant epigenetic marks that are key to a malignant phenotype.
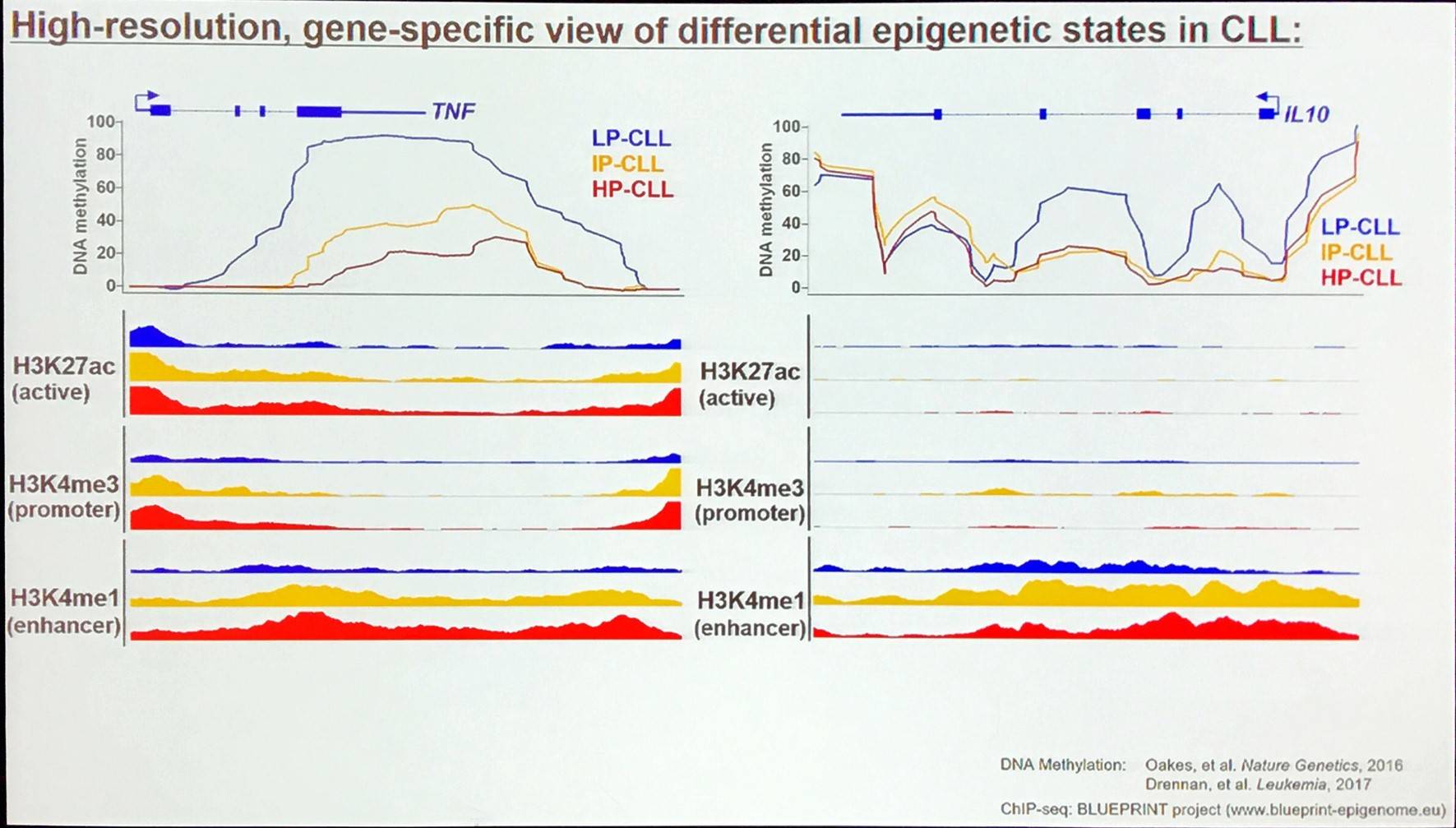
Where do the different epigenetic patterns in CLL come from?
CLL subgroup-specific methylation states are mirrored in normal B-cell subsets. A huge amount of DNA methylation takes place during maturation of B-cells and DNA methylation programming is associated with numerous annotated pathways:
|
Members of the BCR signaling pathway |
8.62 |
|---|---|
|
Genes related to PIP3 signaling in B-lymphocytes |
6.21 |
|
Chronic Myeloid Leukemia |
5.95 |
|
Gene regulation by peroxisome proliferators via PPARα |
5.93 |
|
B-Cell Antigen Receptor |
5.56 |
|
Trefoil Factors initiate mucosal healing |
4.96 |
|
Role of Erk5 in neuronal survival |
4.92 |
|
B-Cell survival pathway |
4.91 |
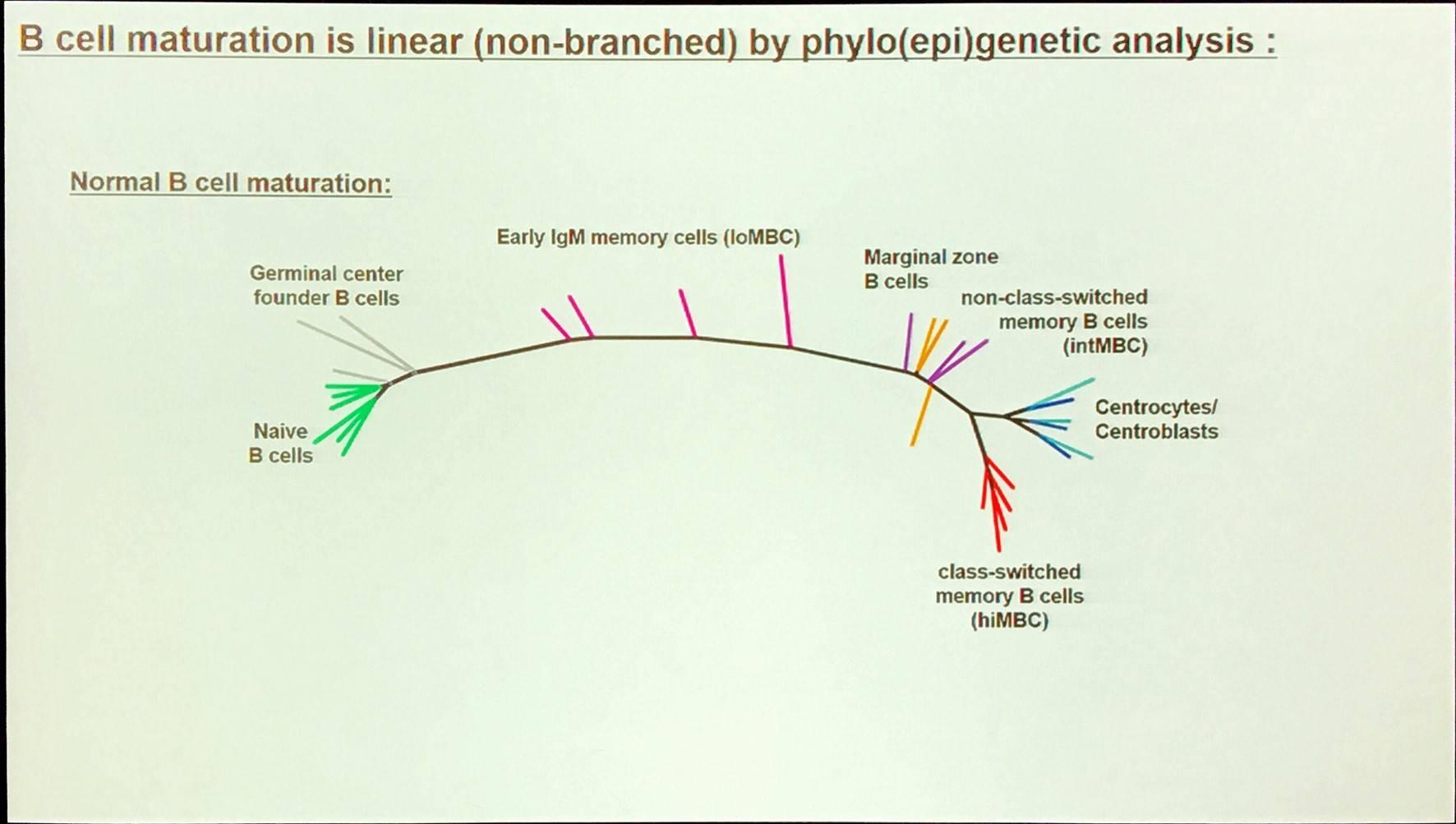
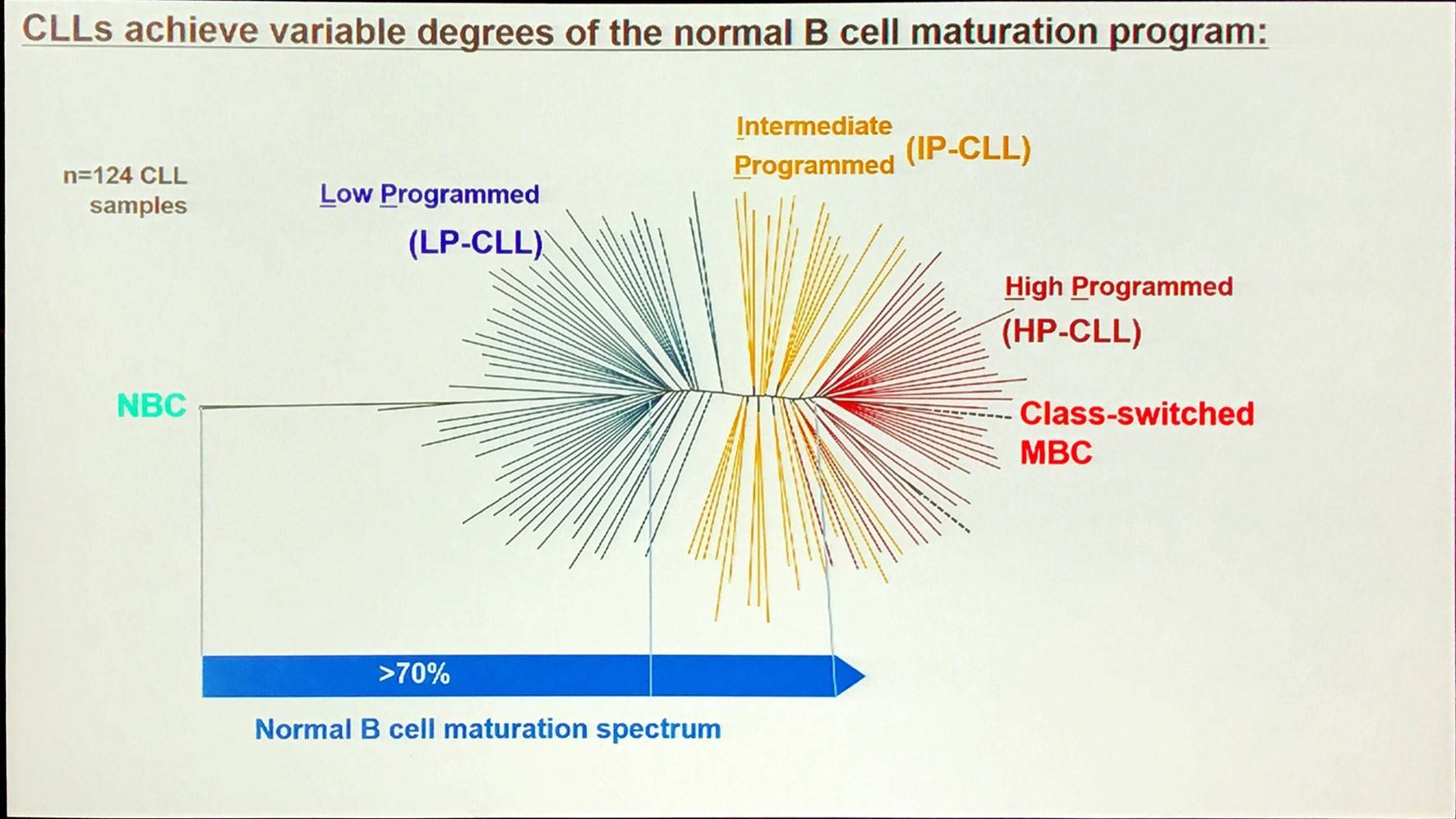
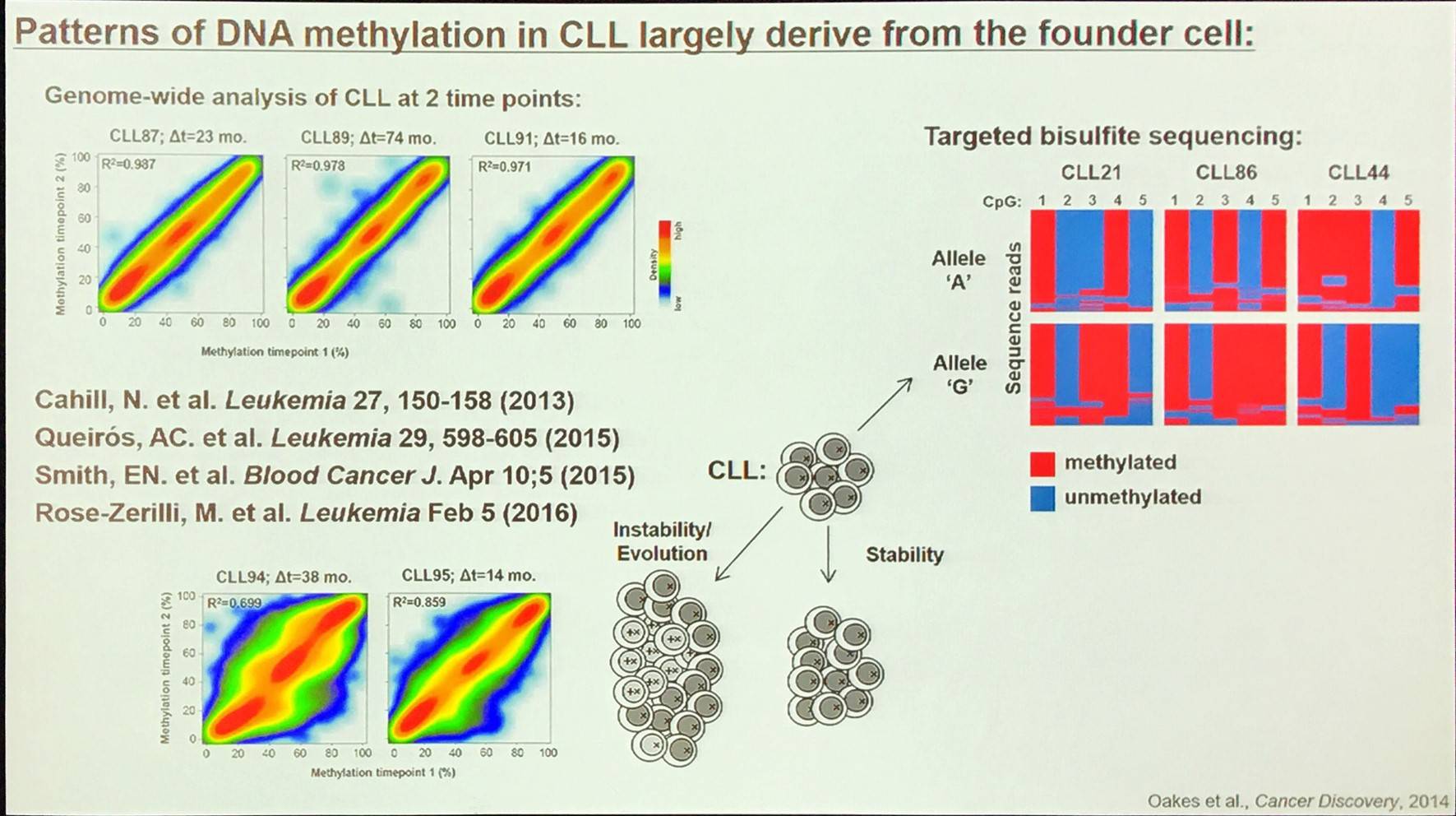
A large proportion of the epigenetic patterns observed in CLL derives from the epigenetic status of the founder cell.
Additionally, aberrant DNA methylation programming of Low Programmed (LP)-CLL involves excess programming of NFAT and EGR as well as reduced programming of EBF and AP-1, unbalancing the normal B-cell epigenetic program (Oakes et al. 2016). CLL-specific methylation may involve aberrant BCR signaling.
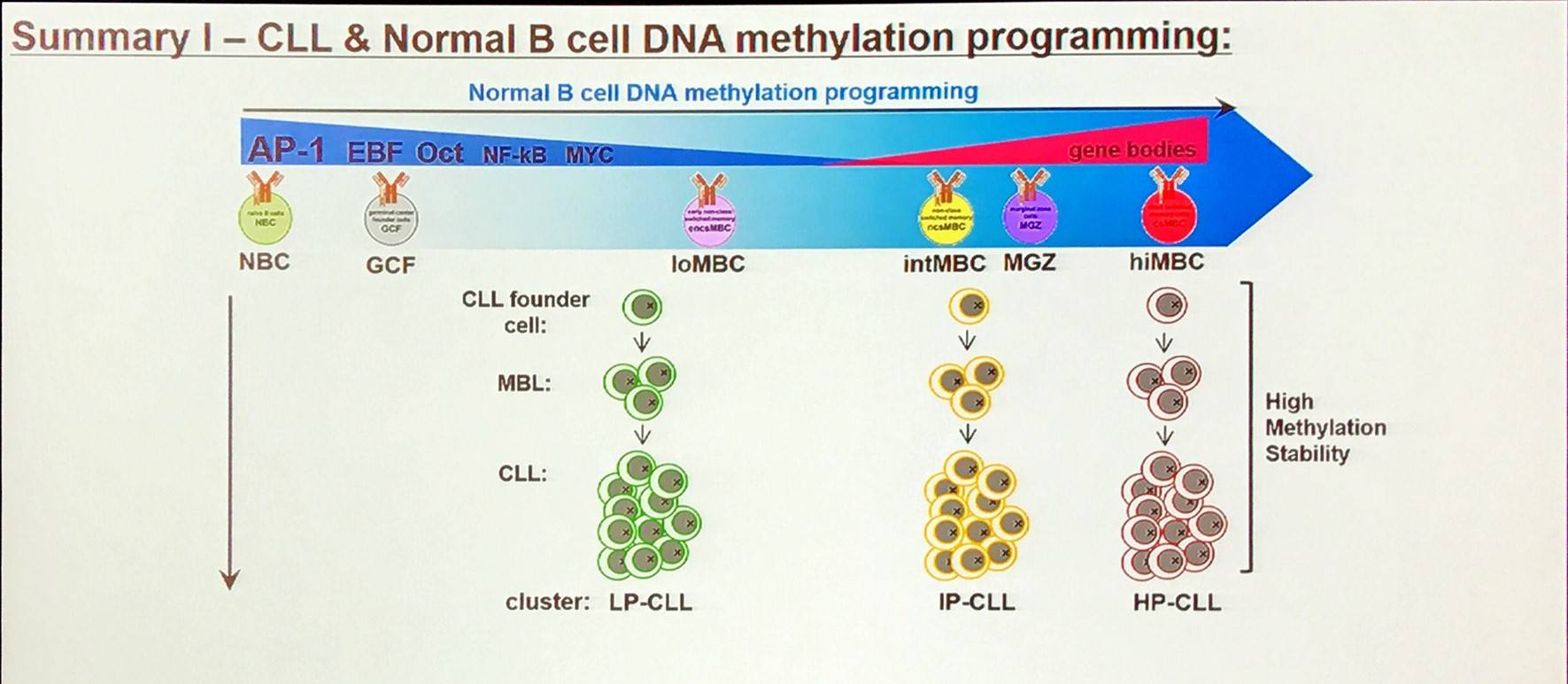
Overall, in CLL, the majority of methylation marks relate to normal B-cell maturation. Moreover, CLL methylation pattern is closer to memory than naïve B-cells. However, LP-CLL selectively maintain naïve B-cell states at specific genes. Lastly, the epitype derives from the maturation state of the cell of origin.
Do epigenetic patterns influence the biology of CLL?
Methylation states control gene expression in CLL (Drennan et al. 2017; Oakes et al. 2016); modulating the expression of hundreds of genes. Additionally, DNA methylation subgroups in CLL has a strong association with clinical outcomes.
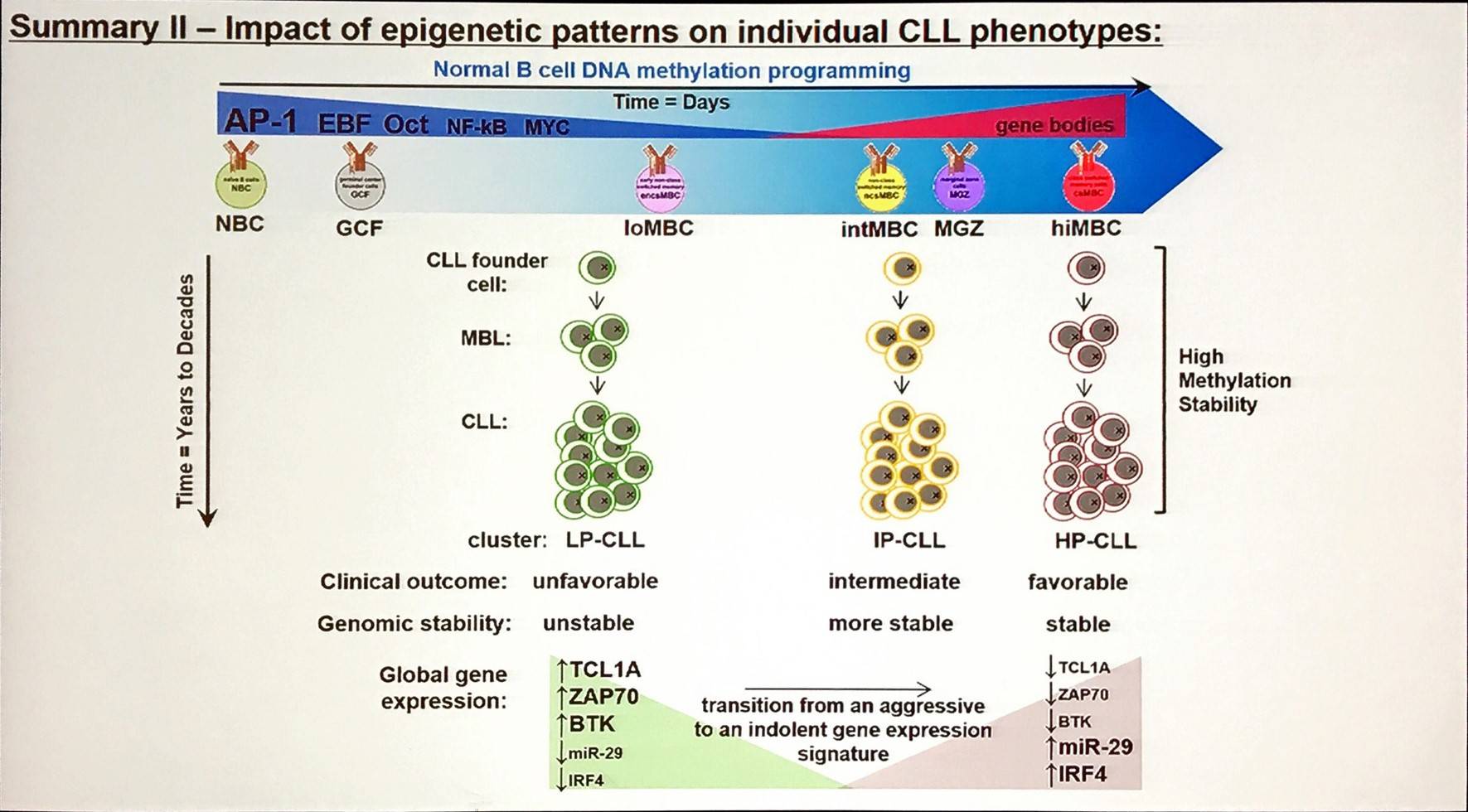
How can we make use of this information?
DNA methylation marks make for powerful biomarkers, for example methylation of ZAP-70:
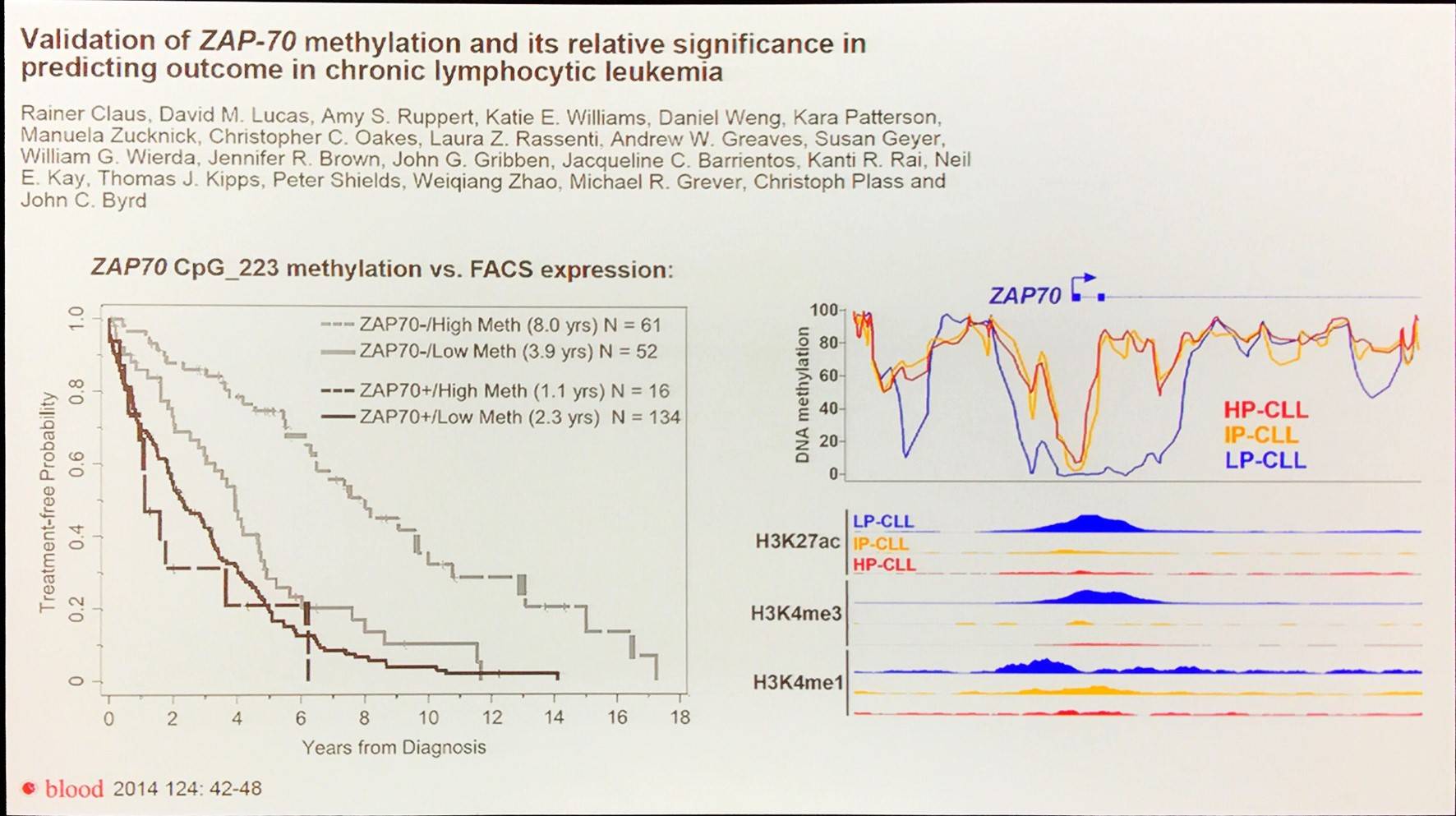
Epigenomic signatures may also reveal the cellular events surrounding the genesis of individual CLLs, identifying potential novel therapeutic vulnerabilities.
- Oakes C. Harnessing epigenetics to further our understanding of CLL ontogeny and clinical behavior. XVII International Workshop on Chronic Lymphocytic Leukemia; 2017 May 12–15; New York, USA.

Understanding your specialty helps us to deliver the most relevant and engaging content.
Please spare a moment to share yours.
Please select or type your specialty
 Thank you
Thank youRelated articles
Newsletter
Subscribe to get the best content related to lymphoma & CLL delivered to your inbox








