All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the Lymphoma Coalition.
The Lymphoma Hub uses cookies on this website. They help us give you the best online experience. By continuing to use our website without changing your cookie settings, you agree to our use of cookies in accordance with our updated Cookie Policy
Introducing

Now you can personalise
your Lymphoma Hub experience!
Bookmark content to read later
Select your specific areas of interest
View content recommended for you
Find out moreThe Lymphoma Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the Lymphoma Hub cannot guarantee the accuracy of translated content. The Lymphoma Hub and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
ICML 2017 | Genotyping of classical Hodgkin Lymphoma using liquid biopsy – Hodgkin Lymphoma Session
Bookmark this article
During the 14th International Conference on Malignant Lymphoma (ICML) held in Lugano, Switzerland, a Hodgkin Lymphoma Session, co-chaired by V. Diehl (University of Cologne, Germany) and A. Pavlovsky (Pavlovsky Center for Hematology, Buenos Aires), took place on Thursday 15th June.
During this session, D. Rossi from the Institute of Oncology Research, Bellinzona, Switzerland, gave a talk on genotyping classical Hodgkin Lymphoma (cHL) using a liquid biopsy (abstract #052).
Thus far, the mutational profile of cHL is poorly characterized and the genetics of refractory disease have not been fully elucidated. Genotyping of tumor tissue is challenging (i.e. tumor representation, FFPE) and interim PET is neither 100% sensitive or specific for defining ultimate patient outcome.
Liquid biopsy is a source of tumor DNA for Lymphoma genotyping. Plasma cell free DNA is easily accessible, allows for real time monitoring, and has already been tested in DLBCL.
The study presented by Rossi aimed to:
- Provide evidence that plasma cfDNA can be used to track the cHL mutational profile
- Characterize the genetics of newly diagnosed cHL and, for comparative purposes, refractory cHL
The study included plasma cfDNA and germline (g)DNA from 29 patients with treatment naïve and 15 patients with chemo-refractory cHL. Paired gDNA from tumor tissue biopsies was available for 17 patients, including 3 cases for which Reed-Sternberg (RS) enriched areas were macro-dissected. Plasma cfDNA, normal gDNA, and tumor gDNA were subjected to targeted ultra-deep Next Generation Sequencing (NGS) by using CAPP-seq and Illumina platforms (sensitivity of 3x10-3).
Key Highlights:
- Plasma cfNDA genotyping identified non-synonymous somatic mutations in:
- Newly diagnosed cHL: STAT6 (45%), TNFAIP3 (45%), ITPKB (31%), B2M (21%), GNA13 (17%), and XPO1 (10%) among the most recurrent genes
- Refractory cHL: GNA13 (36%), ITPKB (29%), ATM (29%), B2M (21%), STAT6 (21%), KMT2D (21%), XPO1 (21%), TET2 (21%), and TNFAIP3 (14%)
- Mutations of TET2 were enriched in refractory cHL patients versus newly diagnosed cases, indicating they contribute to the chemo-refractory phenotype
- Consistently, genotyping of longitudinal samples disclosed the acquisition of TET2 mutations in one patient with refractory disease
- The majority of mutations discovered in cfDNA were also identified in paired tumor DNA from the tissue biopsy and/or macro-dissected RS cells, confirming their tumor origin
- By pathway analysis, the mutational profile indicated the involvement of PI3K/AKT signaling (49%), cytokine signaling (41%), NF-kB signaling (51%), epigenetics (39%), and immune escape (22%) in cHL
- ITPKB (a negative regulator of PI3K) was specifically mutated in cHL across aggressive B-cell Lymphomas
- In RS cells from wild type cases, the ITPKB protein showed a nucleo-cytoplasmic pattern; in RS cells from mutated cases, ITPKB localized only in the cytoplasm, alluding to the mutations resulting in a change in function and alteration of subcellular localization of the protein
- The L-1236 cHL cell line, that harbors a mutated, truncated ITPKB, was resistant to PI3K inhibitors (RP6530 and AEZS136)
- cHL cell lines harboring a wild type ITPKB (L-540, L-428, KM-H2) maintained sensitivity to these compounds
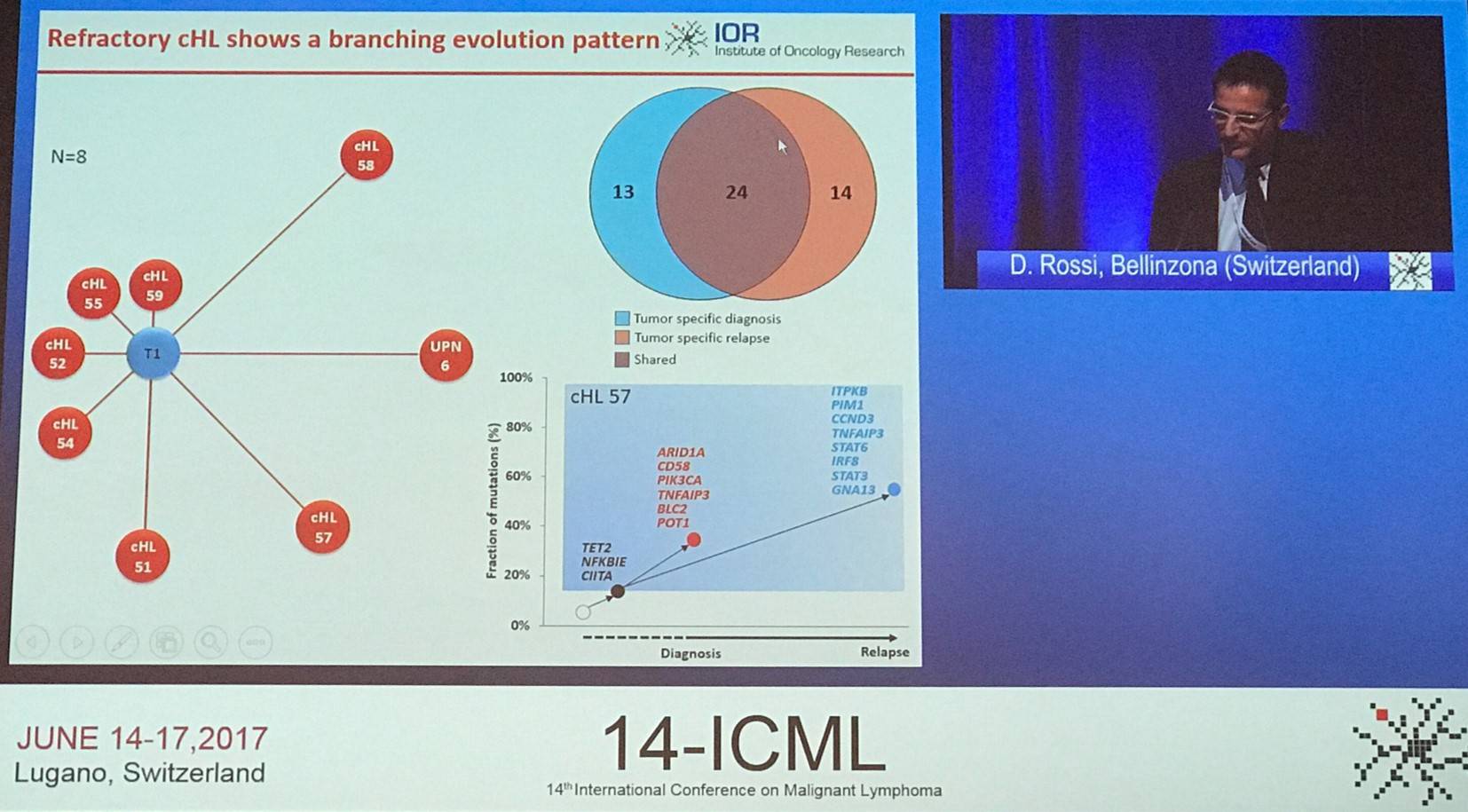
Davide Rossi concluded the talk by stating that cHL can successfully be genotyped using plasma cfDNA. STAT6, TNFAIP3, and ITPKB are frequently (>20%) mutated in cHL. ITPKB mutations (approximately in 30% of cases) associate with pAKT and resistance to PI3K inhibitors. cHL refractoriness does not associate with TP53 mutations but is enriched with TET2 mutations. Clonal evolution in cHL follows a branching pattern. Lastly, dynamic changes in tumor cfDNA may complement imaging in anticipating ABVD outcome.
- Bruscaggin A. et al. Genotyping of Classical Hodgkin Lymphoma on the Liquid Biopsy. Hematological Oncology. 2017 June 07;35(suppl S2):64–5. DOI: 10.1002/hon.2437_51.
More about...

Understanding your specialty helps us to deliver the most relevant and engaging content.
Please spare a moment to share yours.
Please select or type your specialty
 Thank you
Thank youRelated articles
Newsletter
Subscribe to get the best content related to lymphoma & CLL delivered to your inbox








