All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the Lymphoma Coalition.
The Lymphoma Hub uses cookies on this website. They help us give you the best online experience. By continuing to use our website without changing your cookie settings, you agree to our use of cookies in accordance with our updated Cookie Policy
Introducing

Now you can personalise
your Lymphoma Hub experience!
Bookmark content to read later
Select your specific areas of interest
View content recommended for you
Find out moreThe Lymphoma Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the Lymphoma Hub cannot guarantee the accuracy of translated content. The Lymphoma Hub and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
iwCLL 2017 | Insights into inherited susceptibility of CLL
Bookmark this article
On Monday 13th May, the first session during iwCLL 2017 was titled “Unraveling the factors leading to the development of CLL”, and was chaired by Daniel Catovsky (Institute of Cancer Research, London, UK) and Stephan Stilgenbauer (University of Ulm, Germany).
During this session, Richard Houlston, also from the Institute of Cancer Research, London, UK, gave a presentation titled: “Insights into inherited susceptibility to Chronic Lymphocytic Leukemia.”
Houlston began by listing the risk factors for CLL, which include age older than 65 years and being male. Currently, no environmental or lifestyle factors have been found to contribute to risk of CLL.
The talk then discussed if family history is a risk factor for CLL.
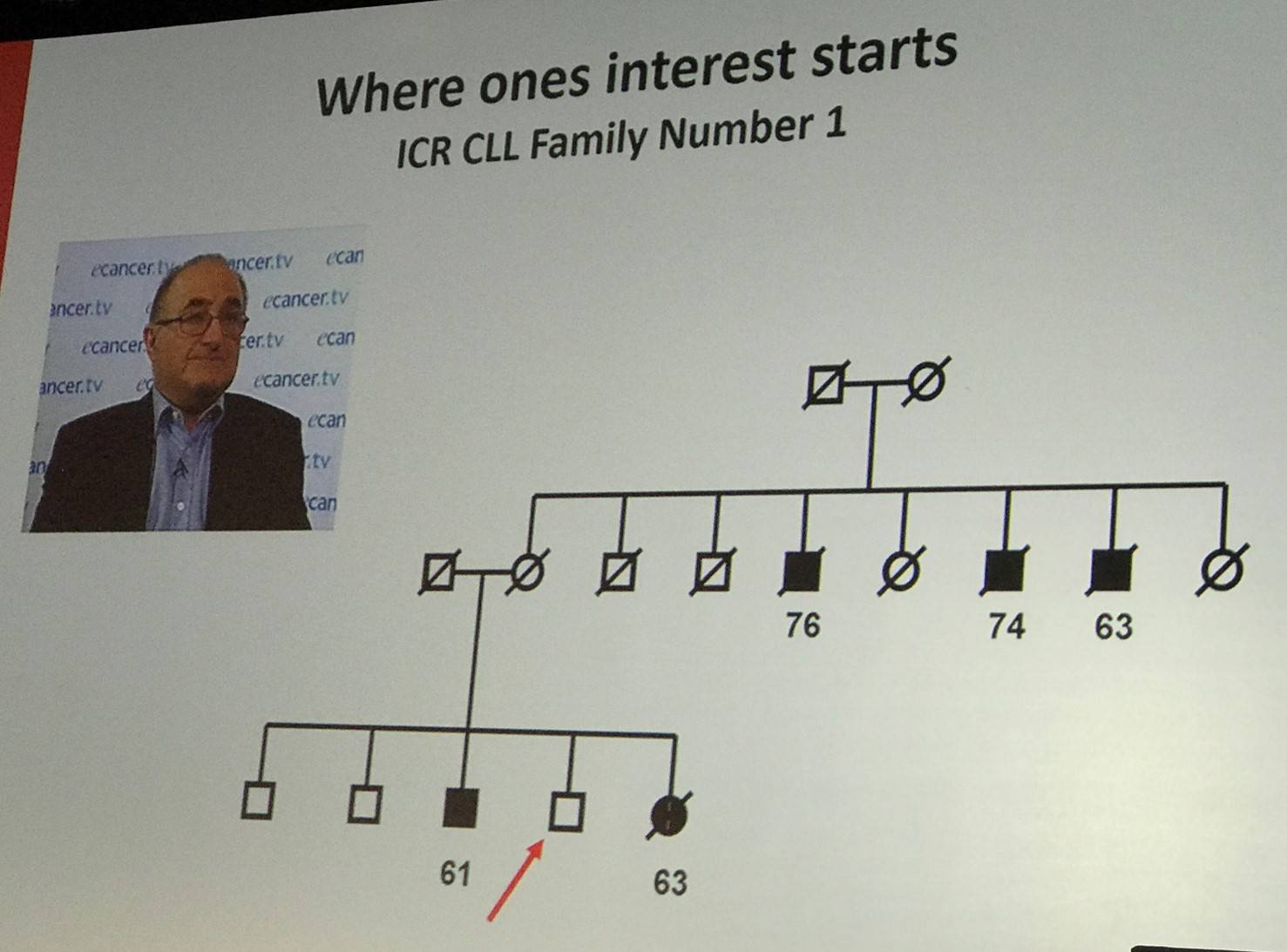
Richard Houlston stated that considering family history as a risk factor is not a new concept. In 1947, Aage Videbaek reported familial clustering of leukemia. More recently, in 2004, L.R. Goldin et al. found that relatives of cases were at significantly increased risk for CLL (7.52-fold compared to relatives of normal controls). Also, data from Gabrielle S. Sellick et al. was presented.
The talk then turned to discussing candidate gene studies. Numerous candidates have been considered including genes of the immune response (P2X7, TNF), carcinogenic metabolism (GSTM1, CYP1A1, NAT2), DNA repair (XRCC2), and methylation (MTHRF). However, between 1981 and 2015, 94 candidate gene studies have been published assessing over 100 genes, but no robust associations have been found.
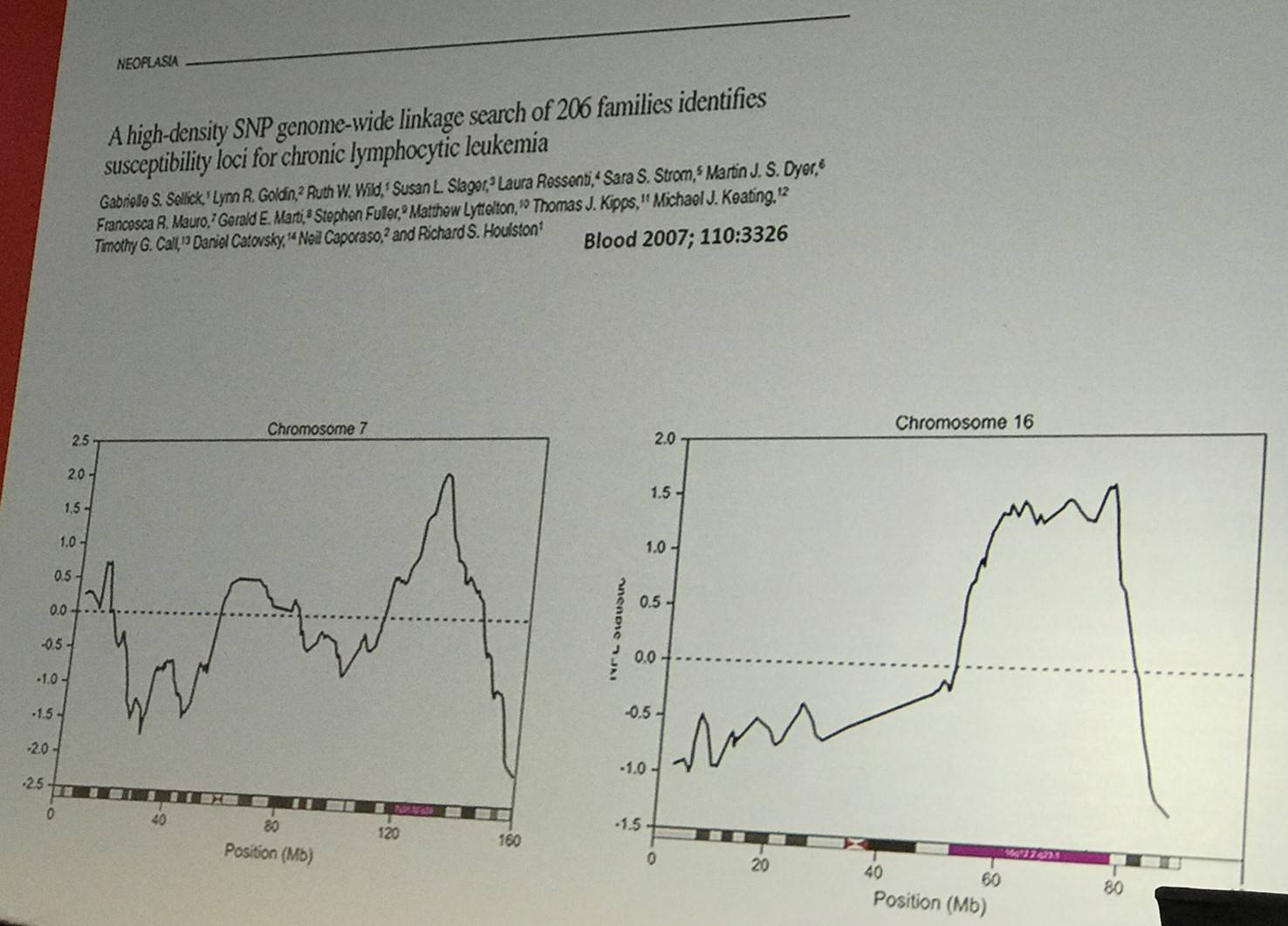
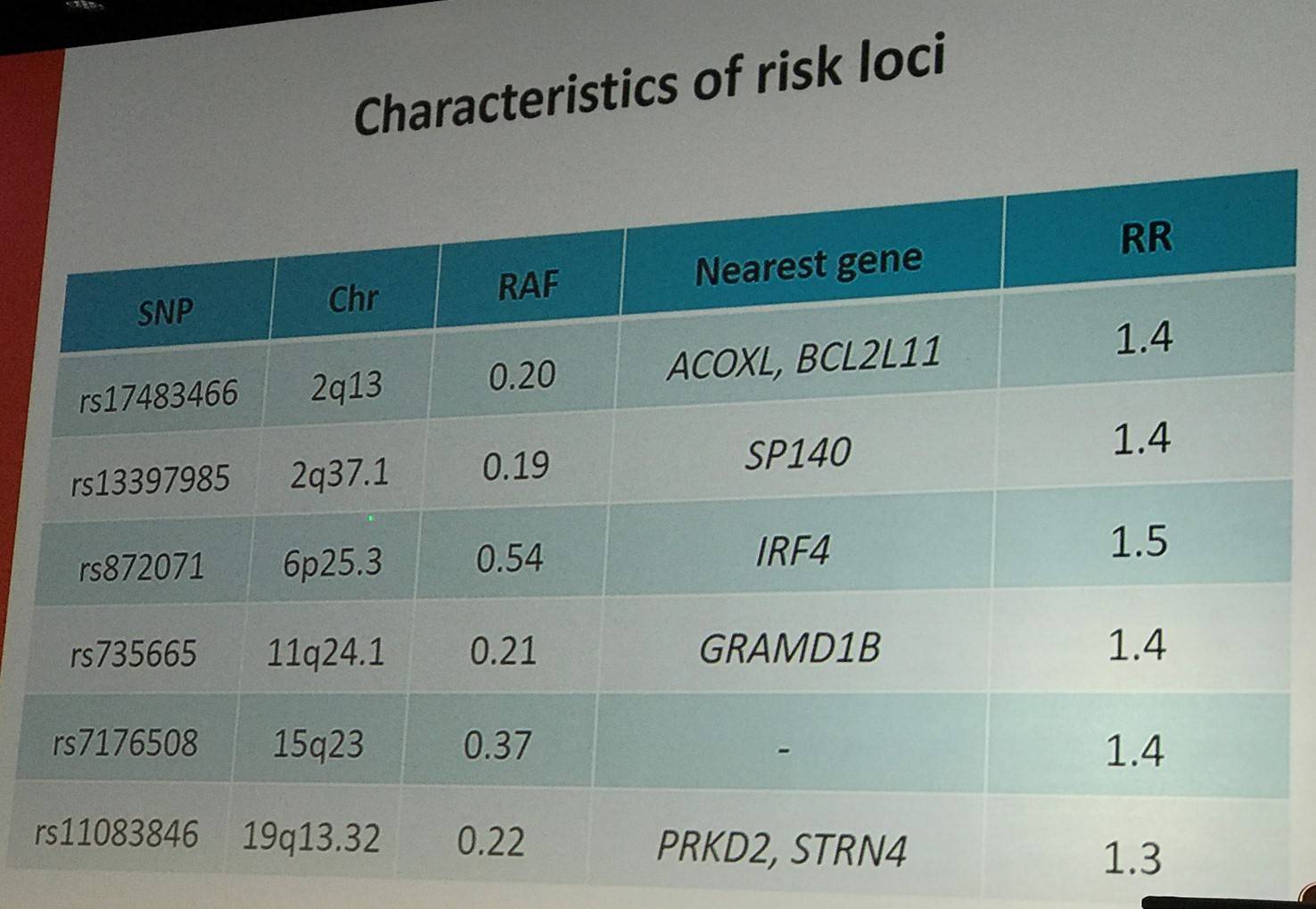
Despite this, we have had more success with Genome-Wide Association Studies (GWAS), which has identified numerous susceptibility loci at P < 10-8. Richard Houlton went on to outline the characteristics of some of these risk loci (slide below). It has also been found that risk loci show enrichment of open chromatin.
The talk then focused in detail on the 15q15.1 risk locus:
- Rs539846 is a causal Single Nucleotide Polymorphism (SNP), BMF is the target gene
- SNP modulates enhancer activity, disrupts RELA binding, and alters BMF expression
- BMF is a BH3-only protein; BH3-mimetics are now novel agents to treat CLL
Houlston then discussed chromosome 8q24. This area displays multiple interactions with Myc and associations have been found in this area of the genome for CLL, Hodgkin Lymphoma, and Myeloma.
The topic of the talk then turned to determining the contribution of polygenic inheritance to CLL:
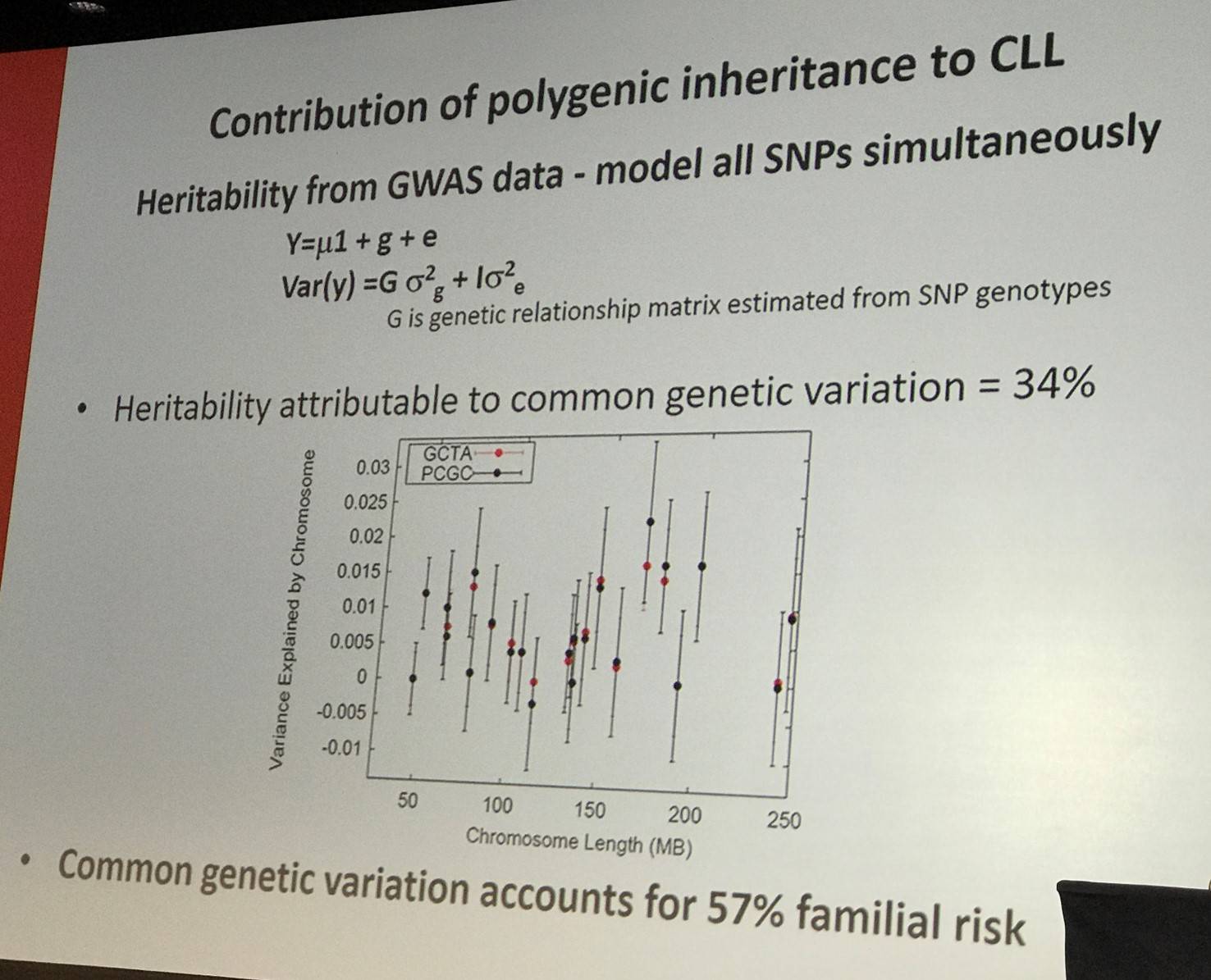
Whole-exome sequencing has been carried out aiming to identify familial CLL genes. In 66 families, with 141 cases of CLL, Loss-of-Function (LOF) mutations were identified, prioritizing genes with a known role in cancer and excluding variants seen in more than 1 of 1,600 healthy 1958BC controls. It was found that there is a 3.6-fold increase risk of CLL with POT1 p.Gln376Arg (rs17246404). Previously, it has been reported that 3.5% of CLL cases carry somatic POT1 mutations (Ramsay et al., 2013). POT1 is a critical factor in maintaining telomere integrity; it is a component of the shelterin complex, which binds directly to DNA. Previously, POT1 mutation has been associated with other cancers such as Cutaneous Malignant Melanoma (Jianxin Shi et al.) and Familial Glioma (Bainbridge et al.).
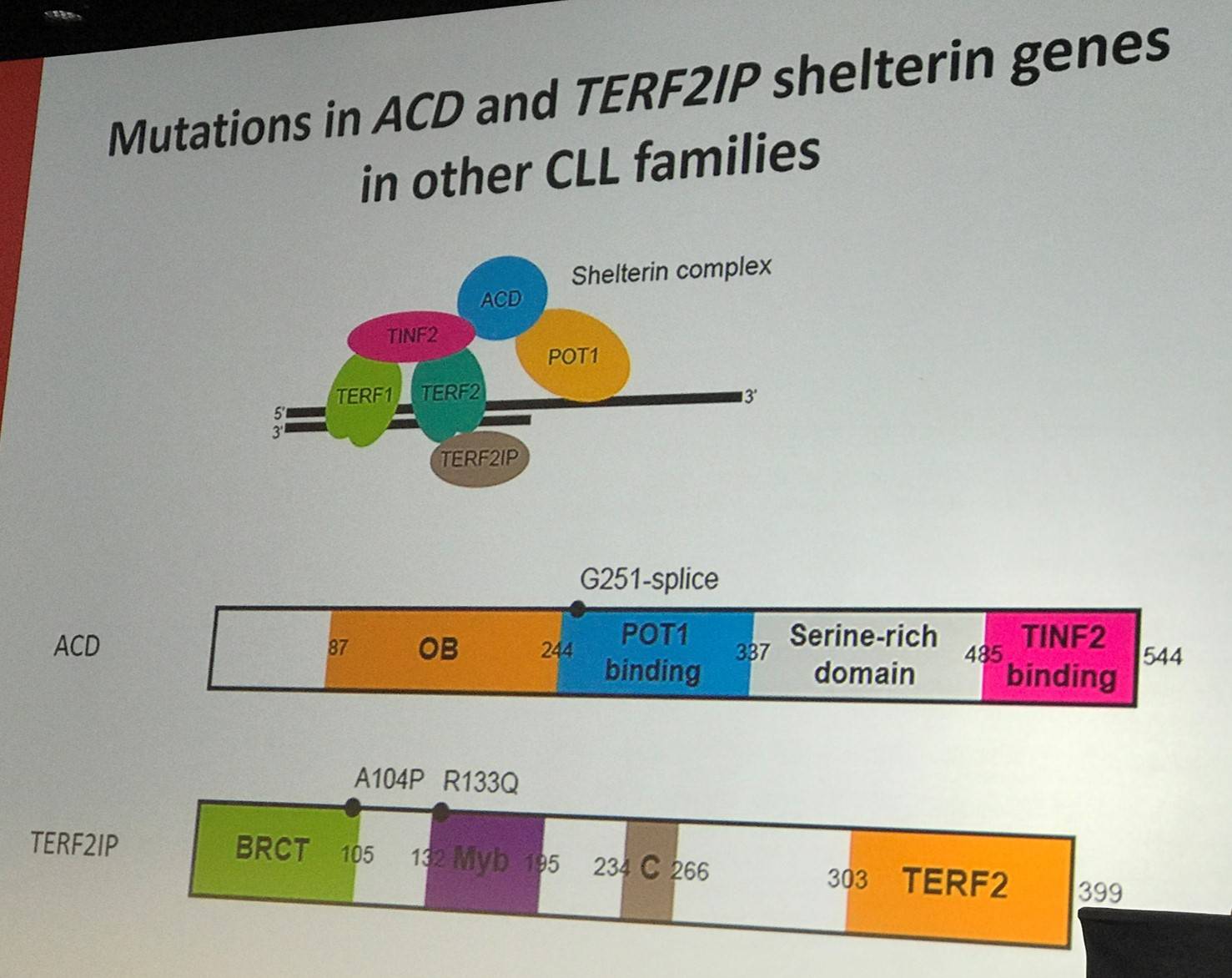
Richard Houlston concluded his talk by stating that there is direct evidence supporting the inherited susceptibility of CLL, and there are a range of alleles and effect sizes that impact on risk. GWAS has been the dominant type of study in recent years, but carries challenges when trying to interpret the findings and associations in terms of biology. Lastly, higher-impact mutations are beginning to be revealed.
- Houlston R. Insights into inherited susceptibility to Chronic Lymphocytic Leukemia. XVII International Workshop on Chronic Lymphocytic Leukemia; 2017 May 12–15; New York, USA.

Understanding your specialty helps us to deliver the most relevant and engaging content.
Please spare a moment to share yours.
Please select or type your specialty
 Thank you
Thank youRelated articles
Newsletter
Subscribe to get the best content related to lymphoma & CLL delivered to your inbox








